Continuing explication of the motions submitted on January 9th to the U.S. Patent and Trademark Office Patent Trial and Appeal Board in Interference No. 106155 between Senior Party The Broad Institute, Harvard University, and the Massachusetts Institute of Technology (collectively, "Broad") and Junior Party the University of California/Berkeley, the University of Vienna, and Emmanuelle Charpentier (collectively, "CVC") is CVC's Motion No. 2 in Opposition to the Broad's Substantive Motion No. 2 to Substitute the Count.
To recap, the Broad's Substantive Motion No. 2 seeks to have the Board substitute the Count in the interference. Count 1 of the interference as declared is:
An engineered, programmable, non-naturally occurring Type II CRISPR-Cas system comprising a Cas9 protein and at least one guide RNA that targets and hybridizes to a target sequence of a DNA molecule in a eukaryotic cell, wherein the DNA molecule encodes and the eukaryotic cell expresses at least one gene product and the Cas9 protein cleaves the DNA molecules, whereby expression of the at least one gene product is altered; and, wherein the Cas9 protein and the guide RNA do not naturally occur together, wherein the guide RNAs comprise a guide sequence fused to a tracr sequence.
or
A eukaryotic cell comprising a target DNA molecule and an engineered and/or non-naturally occurring Type II Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-— CRISPR associated (Cas) (CRISPR-Cas) system comprising
a) a Cas9 protein, or a nucleic acid comprising a nucleotide sequence encoding said Cas9 protein; and
b) a single molecule DNA-targeting RNA, or a nucleic acid comprising a nucleotide sequence encoding said single molecule DNA-targeting RNA; wherein the single molecule DNA-targeting RNA comprises:
i) a targeter-RNA that is capable of hybridizing with a target sequence in the target DNA molecule, and
ii) an activator-RNA that is capable of hybridizing with the targeter-RNA to form a double-stranded RNA duplex of a protein- binding segment,
wherein the activator-RNA and the targeter-RNA are covalently linked to one another with intervening nucleotides; and
wherein the single molecule DNA-targeting RNA is capable of forming a complex with the Cas9 protein, thereby targeting the Cas9 protein to the target DNA molecule, whereby said system is capable of cleaving or editing the target DNA molecule or modulating transcription of at least one gene encoded by the target DNA molecule.
The Broad's proposed Count 2 is:
A method, in a eukaryotic cell, of cleaving or editing a target DNA molecule or modulating transcription of at least one gene encoded by the target DNA molecule, the method comprising:
contacting, in a eukaryotic cell, a target DNA molecule having a target sequence with an engineered and/or non-naturally-occurring Type II Clustered Regularly lnterspaced Short Palindromic Repeats (CRISPR)-CRISPR associated Cas) (CRISPR-Cas) system comprising:
a) a Cas9 protein, and
b) RNA comprising
i) a targeter-RNA that is capable of hybridizing with the target sequence of the DNA molecule or a first RNA comprising (A) a first sequence capable of hybridizing with the target sequence of the DNA molecule and (B) a second sequence; and
ii) an activator-RNA that is capable of hybridizing to the targeter-RNA to form an RNA duplex in the eukaryotic cell or a second RNA comprising a tracr sequence that is capable of hybridizing to the second sequence to form an RNA duplex in the eukaryotic cell,
wherein, in the eukaryotic cell, the targeter-RNA or the first sequence directs the Cas9 protein to the target sequence and the DNA molecule is cleaved or edited or at least one product of the DNA molecule is altered.
The distinction the Broad makes is between embodiments of CRISPR methods that are limited to "single-molecule guide RNA" (aka "fused" or "covalently linked" species), versus embodiments that encompass single-molecule and "dual molecule" species (wherein in the latter versions, the "targeter-RNA" and "activator-RNA" as recited in the proposed Count are not covalently linked). The Broad argues that this Count should be adopted by the Board because it "properly describes the full scope of the interfering subject matter between the parties because both parties have involved claims that are generic, non-limited RNA claims." The brief also argues that proposed Count 2 "sets the correct scope of admissible proofs [i.e., their own] for the breakthrough invention described by the generic claims at issue in these proceedings—the successful adaption of CRISPR-Cas9 systems for use in eukaryotic environments," which The Broad contends current Court 1 (in either alternative) does not.
The brief also requests that the Broad be accorded benefit to its provisional application U.S. Serial No. 61/736,527, and that CVC not be accorded benefit to any of its earlier applications (which just maintains the status quo requiring CVC to move to be accorded benefit), because CVC's applications-in-interference were not accorded benefit when this interference was declared.
The Broad's argument in support of its motion is that Count 1 is too narrow for encompassing just a subset of the parties' involved claims. In particular, the brief asserts that most of the Broad's involved clams encompass "non-limited" RNA systems and methods. Similarly, the brief argues that CVC itself has many claims directed to non-limited RNA systems and methods and has entire applications that do not recite claims to non-limited RNA systems and methods. The Broad asserts that Count 1 does not permit the Broad to rely on its earliest and best proofs of invention, which the brief states is "plainly unfair." This unfairness would preclude the Broad from establishing what the brief terms "the fundamental breakthrough - the invention of use of CRISPR in eukaryotic cells" (emphasis in brief). Failing to substitute the Count would instead improperly focus the priority question on who invented the single molecule modification. Colorfully, the brief declares that "[a]llowing the interference to proceed with Count 1 would permit the (single molecule RNA) tail to wag the (breakthrough use of CRISPR in eukaryotic cells) dog."
The claims include such generic species that encompass both single-molecule and dual molecule embodiments, according to the brief, but the breakthrough invention was adapting CRISPR-Cas9 to be used in eukaryotic cells, an argument the Broad has used consistently here and in the earlier '048 interference to distinguish its claims from CVC's. The brief notes that the first disclosure of this use was in a scientific journal article by some of the Broad's named inventors that "has become by far the most frequently cited CRISPR publication" as evidence of the groundbreaking nature of the invention. Also cited (as a reminder to the Board of its earlier decision, the basis thereof and the Federal Circuit's affirmance of it) is the Board's earlier decision that there was no reasonable expectation of adapting CRISPR for use in eukaryotic cells as further evidence of the breakthrough nature of (as asserted herein) the Broad's invention.
CVC's brief begins by characterizing the Broad's argument that Count 1 of the interference as declared "potentially' exclude Broad's best proofs relating to dual-guide (i.e., dual-molecule) experiments" is a "pretext," because, inter alia, Proposed Count 2 "goes far beyond converting Count 1 into a generic-guide count." Instead, according to CVC, "it transforms Count 1 into a method so broad that it no longer requires formation of the DNA-targeting complex that includes crRNA, tracrRNA, and Cas9." In addition, according to CVC, Proposed Count 2 does not require that the CRISPR-Cas9 complex even have an effect on the target DNA; rather, it recites that "'a product of the DNA' is altered in some unspecified way" (emphasis in brief), which could include (according to CVC) "alterations to RNA or protein caused by processes that are unrelated to the activity of CRISPR-Cas9" including contamination. And the changes the Broad has effected in Proposed Count 2 "have nothing to do with whether the RNA limitation is single-molecule or generic, Broad's only purported reason for needing a new count."
CVC further argues that the Broad's motion is contrary to the provisions of precedential Board decision, Louis v. Okada, 59 U.S.P.Q.2d 1073 (B.P.A.I. 2001). Under Louis, a party must satisfy a three-prong test: "'(1) should make a proffer of the party's best proofs, (2) show that such best proofs indeed lie outside of the scope of the current count, and (3) further show that the proposed new count is not excessively broad with respect to what the party needs for its best proofs.'" CVC's position (explicated in the brief) is that the Broad failed to provide what Louis required for the "significant alterations" made to Count 1 resulting in Count 2. In addition, CVC argues that the Broad has not shown that Count 1 excludes their best proofs, equivocating in using characterizations like its best proofs only potentially fall outside the scope of Count 1. And CVC alleges that the Broad's representations to the Board regarding its best proofs including dual-guide experiments are contradictory to ("flatly contradicts") positions the Broad has taken in ex parte prosecution. And CVC characterizes these prior statements as admissions that preclude the Broad from advancing these arguments in this interference.
In arguing the specifics of what CVC alleges are the Broad's deficiencies in satisfying the Louis standards, CVC quotes the case to the effect that there must be "'a genuine need to change the count, and not simply cause a change for change's sake'" that cannot be supplied outside the brief supporting the motion under 37 C.F.R. § 41.121(b) and 37 C.F.R. § 41.208(b). The changes in Proposed Count 2 "significantly alter the scope of Count 1 in ways that go far beyond encompassing dual-molecule CRISPR-Cas9." The brief provides a table:
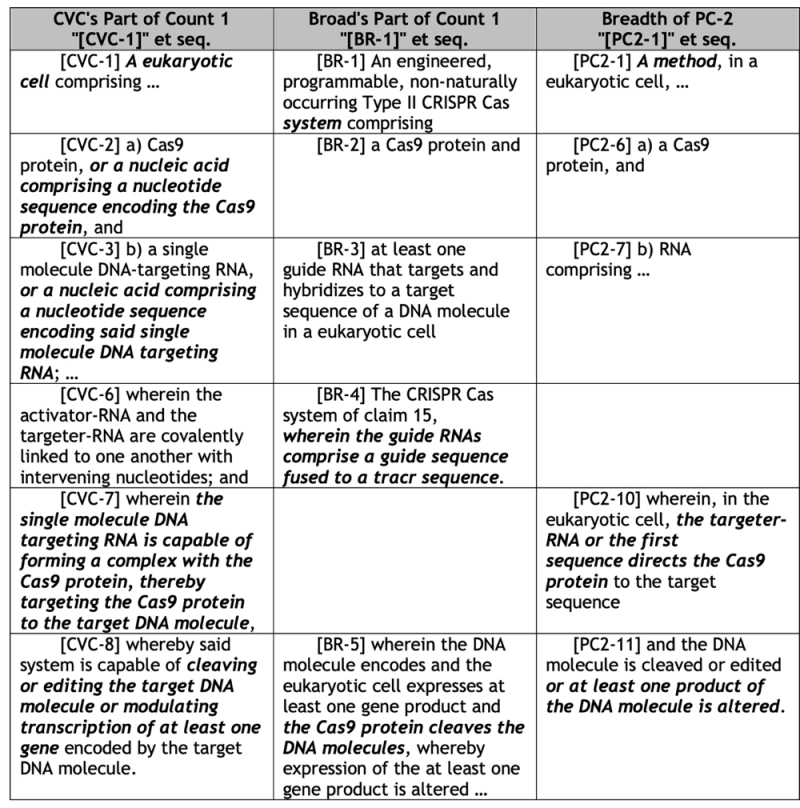 The brief summarizes these unnecessary changes as"
The brief summarizes these unnecessary changes as"
• "first, Broad has inexplicably eliminated structural and functional limitations that specify the formation of the three-component DNA-targeting complex that includes crRNA, tracrRNA, and Cas9."
• "Second, Broad has inexplicably eliminated the requirement that this complex have activity with effects at the DNA level (e.g., cleaving or editing or modulating transcription of DNA). Rather, Proposed Count 2 encompasses merely altering a "product of the DNA molecule" in unspecified ways. Problematically, this breadth includes alterations to downstream products of DNA, such as RNA and protein, that have nothing to do with the activity of the CRISPR-Cas9 system."
• "Third, Broad has inexplicably converted Count 1 from a 'cell' or 'system' to a 'method.'"
• "Fourth, Broad has inexplicably eliminated the alternative language in CVC's part of Count 1 reciting 'or a nucleic acid comprising a nucleotide sequence comprising . . . .'"
CC's brief characterizes as "thread-bare" the Broad's justifications related to its best proofs including use of dual-molecule guide RNA for practicing CRISPR in eukaryotic cells, and argues that the Broad's experts "turned a blind eye" the these differences between Count 1 and Proposed Count 2, "testifying that the only difference they considered was with respect to the single-molecule or generic format of the RNA." According to CVC's brief, the Broad could have achieves its purported "best proofs" goal by proposing a McKelvey Count reciting Claim 15 of the '359 patent:
An engineered, programmable, non-naturally occurring Type II CRISPR-Cas system comprising a Cas9 protein and at least one guide RNA that targets and hybridizes to a target sequence of a DNA molecule in a eukaryotic cell, wherein the DNA molecule encodes and the eukaryotic cell expresses at least one gene product and the Cas9 protein cleaves the DNA molecules, whereby expression of the at least one gene product is altered; and, wherein the Cas9 protein and the guide RNA do not naturally occur together.
CVC also argues that Proposed Count 2 "does not specify that the DNA-targeting complex contains: (a) the first RNA and the second RNA; or (b) the targeter-RNA and the activator-RNA" (emphasis in brief), instead requiring only that "the targeter-RNA or the first sequence directs the Cas9 protein to the target sequence," an interpretation that CVC demonstrates Broad's experts did not dispute despite the reality that all three components are necessary for the DNA targeting complex. And CVC points out that Proposed Count 2 is consistent with the understanding in the art prior to publication of the Jinek 2012 reference that corresponds to disclosure of CVC's earliest priority document (A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity, Science 337: 816-21).
CVC reiterates its argument that Proposed Count 2 does not require CRISPR-Cas9 to exhibit DBA-altering effects due to its excessive breadth, again affirmed (or at least not effectively rebutted" by the Broad's experts. Moreover Proposed Count 2 changes the count from being directed to a eukaryotic cell or a CRISPR system to a method, which CVC characterizes as "change for change's sake" and that "without justification" Count 2 deletes limitations related to "a) Cas9 protein, or a nucleic acid comprising a nucleotide sequence encoding the Cas9 protein," [CVC-2], and "b) a single molecule DNA-targeting RNA, or a nucleic acid comprising a nucleotide sequence encoding said single molecule DNA targeting RNA," [CVC-3]. Citing Louis, CVC argues that the Broad has not shown a "compelling reason" for the proposed changes making Proposed Count 2 broader than Count, and that the proposed Count is "broader than the parties' common invention," citing Conservolite, Inc. v. Widmayer, 21 F.3d 1098, 1104 (Fed. Cir. 1994).
Finally, CVC argues that the Broad's claims in interference all recite "single-molecule RNA" embodiments of CRISPR, which defines the "common interfering subject matter" and thus rebut the Broad's arguments regarding its best proofs for dual-molecule RNA embodiments. The brief accuses the Broad of equivocating regarding its best proofs, also contrary to the requirements in Louis and that the Broad's "alleged best proofs" do not justify substitution of Count 1 with Proposed Count 2 and deficiencies in the Broad's expert testimony supporting its position. These deficiencies include characterization of the prior art that contradict statements and positions taken by the Broad (according to CVC) during ex parte prosecution which CVC asserts are admissions that prevent the Broad from making these assertions in support of its motion to substitute the Count.
CVC further asserts that the Broad has not shown that Proposed Count 2 is patentable over the prior art, as set forth in another table:
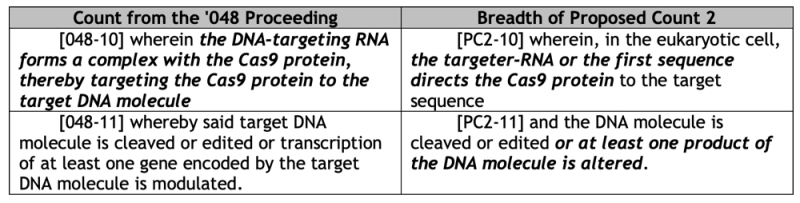
From this CVC concludes "[b]ecause these numerous unexplained broadening alterations raise "a serious question" with respect to the patentability of Proposed Count 2, Broad has not satisfied its burden to address patentability over the art," citing Louis.
As an alternative, CVC argues that, if the Board adopts what the brief calls "a generic-guide count," Count 1 should remain in the interference because it is patentably distinct from Proposed Count 2, supported by the differences in limitations as set forth in the remainder of the brief.