
Growth is one of the defining properties of being biologically alive, and the biology of growth involves both cellular proliferation and differentiation from stem cells to one or a plurality of differentiated cells making up a bodily tissue. Examples range from regeneration of an arm in starfish (and in the related sea cucumber, the ability to propel its gastrointestinal tract to distract a predator) through phylogeny to the capacity of human liver to regrow after resection. Even normal development involves production of new tissues, and in some species both aspects of this biological phenomenon can be appreciated. For example, deer grow and regrow antlers, which can account for 28% of the animal's body weight, and their regrowth rates are so rapid that deer can produce 10 kg or more over 2-3 months. Deer antlers and human bone, while showing very different growth rates (2 cm/day for deer antler, 2 cm/yr for femoral growth during puberty) and other features have in common intramembranous and endochondrial modes of ossification, and this similarity prompted a research group* to undertake a study comparing the pattern of expressed genes in deer antlers and human bone. The results of this study, entitled "Identifying deer antler uhrf1 proliferation and s100a10 mineralization genes using comparative RNA-seq," were published on October 31 in Stem Cell Research and Therapy.
Due to difficulties in elucidating the genes involved in antler production in the past, these researchers initiated their study by isolating reserve mesenchymal (RM) cells from fallow deer and human mesenchymal stem cells (hMSCs) and comparing RNA expression patterns in cultures of both cell types. RM cells were chosen for the study because they expressed robust amounts of alkaline phosphatase (ALP) when contacted with bone morphogenic protein 2 (BMP-2) when assayed using immunofluorescence staining. Cell growth for RM and hMSCs cultures were determined, and RM cells yielded 1.1-4.5 x 105 cells after six day's growth in culture with a doubling time of 17.9-24.7 h and hMSCs yielded 1.2-3.9 x 104 cells after six days in culture (10-fold fewer cells) and a doubling time of 37.8-60.1 h. Differentiation studies showed RM cells differentiated into chondrocytes and osteocytes rather than adipocytes (the predominant differentiated cell type for hESCs). In the presence of BMP-2, RM cells expressed ALP as well as osteocalcin, osteoblast-specific factor 1 and runt-related transcription factor, consistent with the results of the differentiation studies. RM cells were also preferentially stained with Alizarin Red S in mineralization studies (1702.2.ug/mL) compared with hESCs (98.0 ug/mL). These and other comparison of RM and hESCs supported the use of these two cell types for further studies of gene expression related to antler and bone growth in these two species.
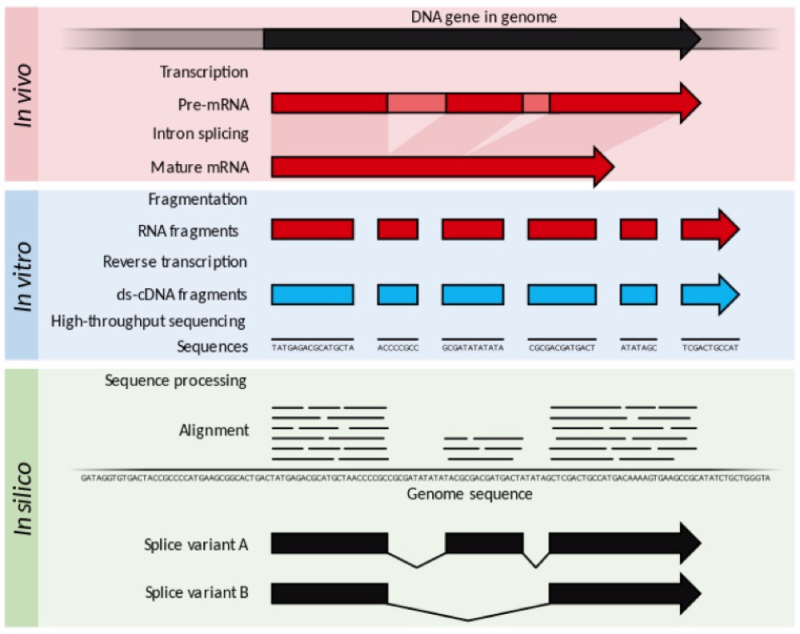
Comparator RM and hMSCs cultures were assayed using RNA-seq methodology, which permits the complement of expressed RNA species in a cell to be determined and compared using a next-generation sequencing (NGS) approach:
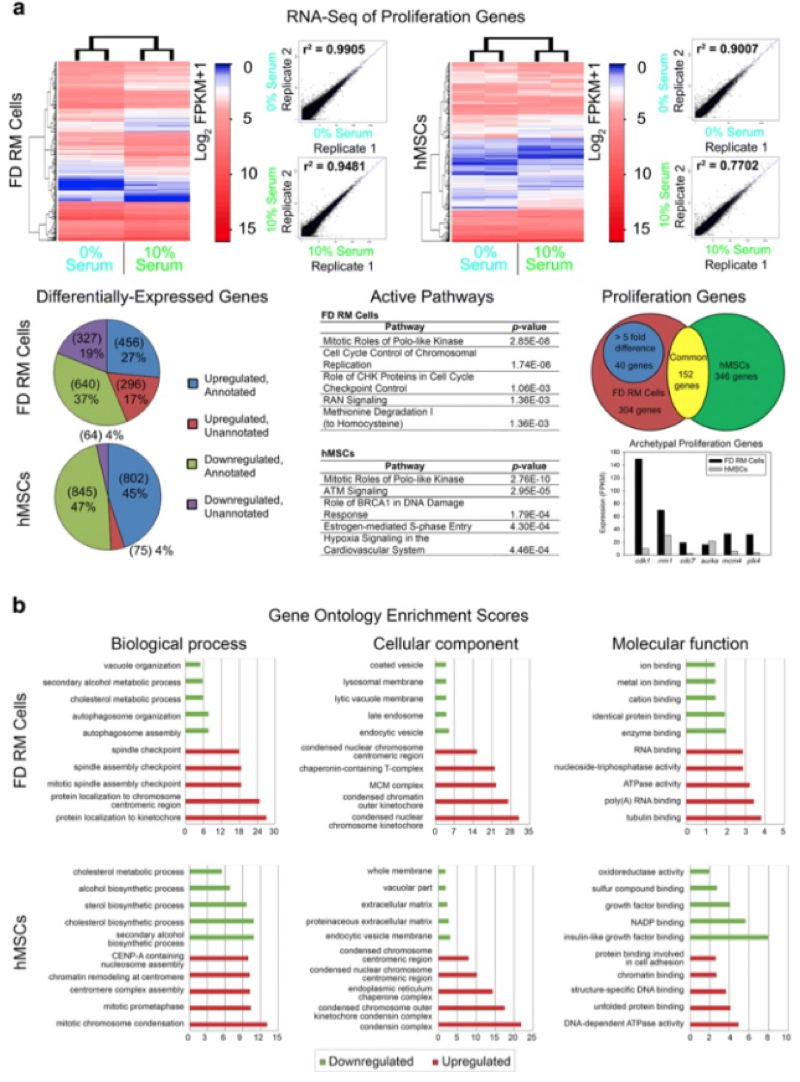
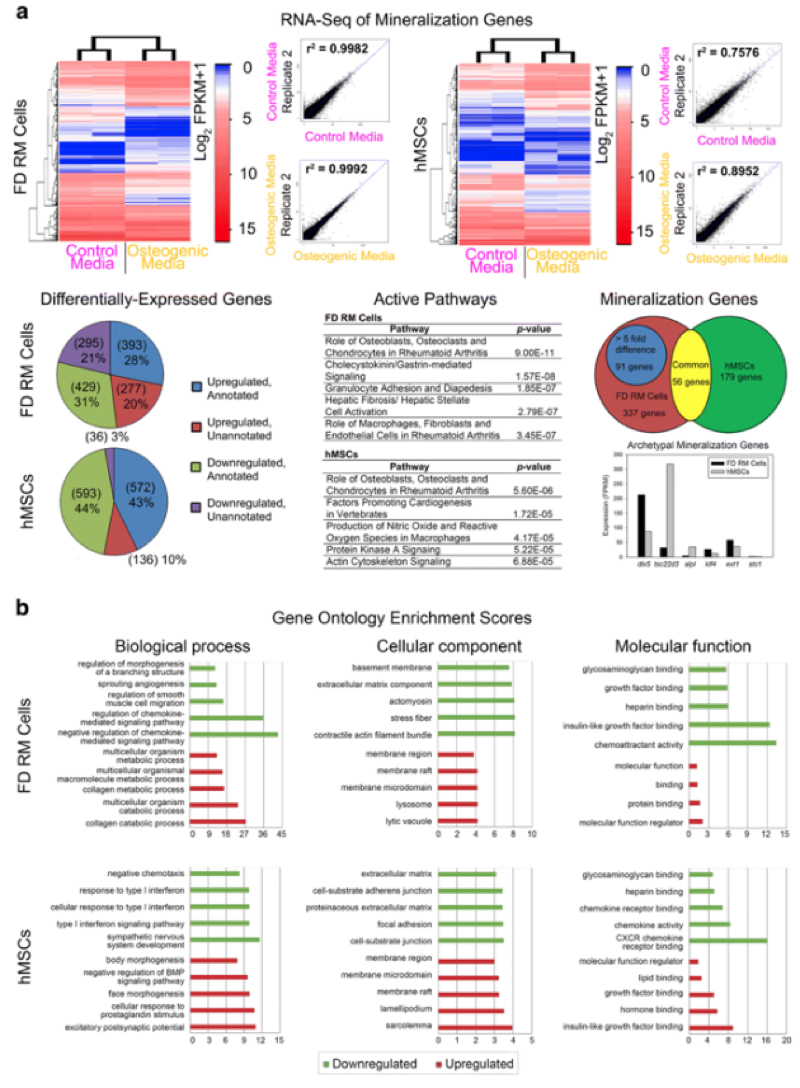
The results of these studies showed similar patterns of activation of proliferation-associated genes in both cell types, including cdk1, rrm1, cdc7, aurka, and plk4. Under conditions expected to promote mineralization both cells types showed "similar activation of osteogenic-associated pathways such as roles of osteoblasts, osteoclasts, and chondrocytes in rheumatoid arthritis as well as the expression of typical osteogenic genes such as dlx5, tsc22d3, alpl, klf4, ext1, and stc1." Overall, these researchers identified 40 proliferation and 91 mineralization candidate genes from these two species:
These results were verified by immunofluorescence, gene over-expression and gene knockdown studies, using FD-derived proliferation candidate gene uhrf1 gene and mineralization candidate gene s100a10. These results were consistent with the purported role in antler growth and regeneration suggested by gene expression patterns identified in in vitro cultured RM cells.
The authors of this study suggest that "[t]he approach developed here may be broadly applied towards studying another biological phenomenon, and the genes identified with this approach will not only advance our understanding of mammalian bone regeneration but also offer promising therapeutic strategies for bone tissue engineering." In addition, they speculate that "a similar comparison strategy can be applied to almost any tissue for identifying the contributions of uniquely expressed genes to a phenomenon of interest." These sentiments are supported by the results set forth in this paper, where a previously intractable biological problem (identification of genes associated with deer antler growth) was successfully addressed using the combination of in vitro biological and RNA-seq approaches set forth herein.
*Dai Fei Elmer Ker, Dan Wang, Rashmi Sharma, Bin Zhang, Ben Passarelli, Norma Nef, Chunyi Li, William Maloney,Stephen Quake and Yunzhi Peter Yang