When I awoke, the dire wolf
Six hundred pounds of sin
Was grinning at my window
All I said was, "Come on in"
But don't murder me, I beg of you
Don't murder me, please, don't murder me
"Dire Wolf," Grateful Dead, Workingman's Dead
The dire wolf (Canis dirus), prototype of the various wolves that were important members of the House Stark family of characters in Game of Thrones, was found uniquely in North America until its extinction in the late Pleistocene (~13,000 years ago). The relationship between this species and the indigenous gray wolf (Canis lupus), the coyote (Canis latrans), and the Asiatic wild dog or dhole (Cuon alpinus) is uncertain, however, based purely on shared morphologic characteristics. Perhaps this "lone wolf" property, and its great size (~68kg/150lbs, despite artistic exaggeration), contributed to its iconic stature in popular culture as diverse as George R.R. Martin and the Grateful Dead, but "mythic" is not too exaggerated a description of this mighty beast in popular culture.
Recently, a diverse and international group of researchers* have explored the relationships between these North American canid species, using both morphological characteristics and genetic comparisons between modern wolves and dire wolf fossils, from both mitochondria and, in a more limited extent genomic DNA (albeit focusing on comparisons of only one gene, COL1). Sites where the more than 700 fossil dire wolf specimens used in these studies were obtained (and their associated academic institutions) were Natural Trap Cave, Wyoming (University of Kansas); Gigantobison Bay, Idaho (Idaho Museum Natural History); Sheridan Pit, Ohio (Cincinnati Museum Center); Guy Wilson Cave, Tennessee (University of Tennessee); American Falls Reservoir, Idaho (Idaho Museum Natural History); and Rancho La Brea Tar Seeps, California (La Brea Tar Pits and Museum), the latter being the predominant site for obtaining dire wolf fossils (100-fold more than gray wolf fossils from this site).
The paper begins by debunking (somewhat) the idea that dire wolves were "sister-species" or conspecific with gray wolves. Although morphology is highly similar between these species as evaluated by various comparative criteria, these authors screened 46 fossil dire wolf specimens dating between 12,900 and 50,000 years ago for mitochondrial DNA (mtDNA) and, at lower complexity, genomic DNA (which is much less well preserved). One of the La Brea Tar Pit specimens provided DNA from type-1 collagen gene (COL1) of sufficient quality for comparison. These experiments suggested that dire wolves were not closely related to either gray wolves, coyotes, African wolves (Canis lupaster) or dogs (Canis familiaris). mtDNA analyses supported these results, that dire wolves formed their own group evolutionarily highly divergent from grey wolves and coyotes, as illustrated by this phylogenetic tree:
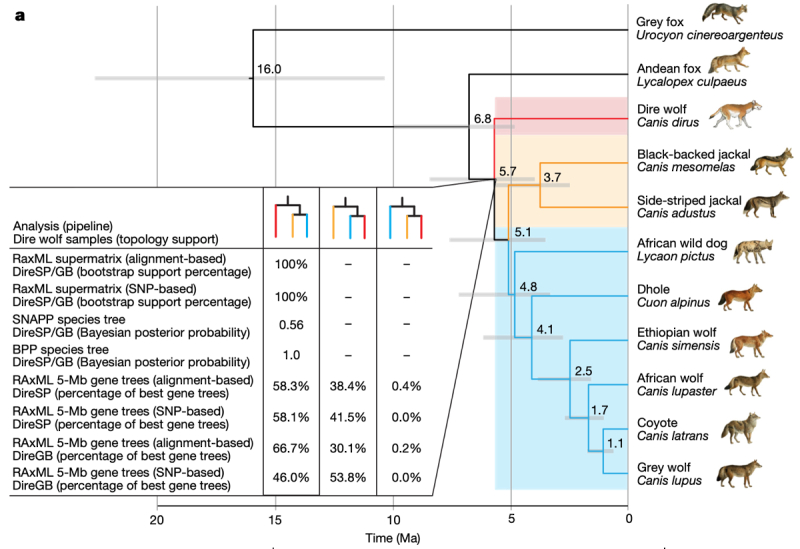
Further, these researchers analyzed nuclear genomic data of the dire wolves together with previously published genomic data from eight extant canids: grey wolf, coyote, African wolf, dhole, Ethiopian wolf (Canis simensis), African wild dog (Lycaon pictus), Andean fox (Lycalopex culpaeus) and grey fox (Urocyon cinereoargenteus—an outgroup). Only the grey wolves, coyotes, dholes, and grey foxes had geographical ranges that overlapped the dire wolf during the Pleistocene prior to its extinction. These analyses confirmed what these scientists characterized as a "distant evolutionary relationship" between the dire wolf and other canid species. Further genomic comparisons indicated that the dire wolf was one branch of the canid lineage (the other two being African jackals and all other canid species), giving the surprising result that grey wolves are more closely related to African wild dogs, Ethiopian wolves, and dholes than they are to dire wolves.
Using these results to provide a "clock" to estimate when dire wolves diverged from other canid species, this paper reports that dire wolves shared a common ancestor with other canid species about 5.7 million years ago (the estimate having a range of 4.0-8.5 million years), with further divergence from African jackals about 5.1 million years ago (3.5-7.6 million years ago). Their genetic analyses also showed no evidence of extensive interbreeding between dire wolves and any other extant North American canid species. There was some evidence of genetic admixture between ancestors of dire wolves and ancestors of grey wolves, coyotes, and dholes about 3 million years ago. These results are consistent with the ready genetic admixture that occurs, inter alia, between modern coyotes and grey wolves. The authors state that, in this context, "our finding of no evidence for gene flow between dire wolves and grey wolves, coyotes or their common ancestor—despite substantial range overlap with dire wolves during the Late Pleistocene—suggests that the common ancestor of grey wolves and coyotes probably evolved in geographical isolation from members of the dire wolf lineage." Given evolution of other canid species, and their status of immigration from Asia, these results suggest to these researchers that the dire wolf arose in North America and may be related to the also-extinct Armbruster's wolf (Canis armbrusteri). Their genetic analyses also suggest to these researchers that dire wolves are properly classified as their own separate genus, Aenocyon, an idea first proposed (not on genetic comparison grounds) in 1918 (see Merriam, J. C., 1918, Note on the systematic position of the wolves of the Canis dirus group. Bull. Dept. Geol. Univ. California 10, 531–33). This genetic and geographic isolation of the dire wolf from other canid species would explain the estimated age of the dire wolf lineage and the evolutionary distance of this species from other canids.
The paper ends with speculation regarding why dire wolves became extinct at the end of the Pleistocene (associated with megafaunal extinctions, i.e., die-offs of animals with weights greater than 100 pounds such as the American lion, the short-faced bear, mammoths, mastodons, ground sloths, and giant beavers) and other canids like coyotes and grey wolves in North America did not, in view of the "overall phenotypic similarities" between these beasts. These speculations include greater morphological plasticity and dietary flexibility in canid species other than the dire wolf, or the ability to interbreed with other canids (which has been shown to permit acquired traits like coat color and enhanced immunity). The researchers suggest that the inability of dire wolves to benefit from such phenotype acquisition through interbreeding might have prevented them from resisting diseases carried by Old World taxa arriving over the Bering land bridge.
While the information is limited (e.g., only five dire wolf genomic DNA samples were sufficiently intact to be assayed), the results from these studies are another example of the power of genetic analysis to "fill in the blanks" from the fossil record to illuminate animal (and human) migration in the Pleistocene era that led to population patterns of animals in the New World in the Holocene (current) Era.
* Department of Archaeology, Durham University, Durham, UK; Australian Centre for Ancient DNA, School of Biological Sciences, University of Adelaide, Adelaide, South Australia, Australia; Department of Ecology and Evolutionary Biology, University of California, Los Angeles, CA; School of Biological and Chemical Sciences, Queen Mary University of London, London, UK; Department of Archaeology, Classics and Egyptology, University of Liverpool, Liverpool, UK; School of Natural Sciences and Psychology, Liverpool John Moores University, Liverpool, UK; The Palaeogenomics & Bio-Archaeology Research Network, Research Laboratory for Archaeology and History of Art, The University of Oxford, Oxford, UK; Department of Anatomy, Des Moines University, Des Moines, IA; Department of Zoology, University of Oxford, Oxford, UK; Department of Anthropology, National Museum of Natural History, Smithsonian Institution, Washington, DC; Center of Excellence in Paleontology & Department of Geosciences, East Tennessee State University, Johnson City, TN; Department of Archaeology, University of Exeter, Exeter, UK; Institute of Archaeology, Russian Academy of Sciences, Moscow, Russia; ARAID Foundation, Instituto Universitario de Investigación en Ciencias Ambientales (IUCA) - Aragosaurus Group, Universidad de Zaragoza, Zaragoza, Spain; Department of Earth Sciences, Natural History Museum, London, UK; Section for Evolutionary Genomics, The GLOBE Institute, University of Copenhagen, Copenhagen, Denmark; Applied Paleoscience, Bothell, WA; Department of Archaeology, University of Sydney, Sydney, New South Wales, Australia; Department of Archaeology, University of Aberdeen, Aberdeen, UK; Department of Archaeology, Simon Fraser University, Burnaby, Canada; Institut des Sciences de l'Evolution - Montpellier, CNRS, Université de Montpellier, IRD, EPHE, Montpellier, France; Laboratoire Evolution & Diversité Biologique, UPS/CNRS/IRD, Université Paul Sabatier, Toulouse, France; Australian Museum Research Institute, Australian Museum, Sydney, New South Wales, Australia; Department of Ecology and Evolutionary Biology, University of California Santa Cruz, Santa Cruz, CA; Institute of Plant and Animal Ecology, Urals Branch of the Russian Academy of Sciences, Yekaterinburg, Russia; Ural Federal University, Yekaterinburg, Russia; Department of Anthropology, Texas A&M University, College Station, TX; Center for Evolution and Medicine, Arizona State University, Tempe, AZ; School of Human Evolution and Social Change, Arizona State University, Tempe, AZ; Halmos College of Arts and Sciences, Nova Southeastern University, Fort Lauderdale, FL; Department of Archaeology, University of York, York, UK; Institute of Systematics and Ecology of Animals, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia; Idaho Museum of Natural History, Idaho State University, Pocatello, ID; Zoological Institute of the Russian Academy of Sciences, St Petersburg, Russia; Sobolev Institute of Geology and Mineralogy, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia; Tomsk State University, Tomsk, Russia; McDonald Institute for Archaeological Research, University of Cambridge, Cambridge, UK; Greenland Institute of Natural Resources, Nuuk, Greenland; NTNU University Museum, Trondheim, Norway; Institute of Human Origins, Arizona State University, Tempe, AZ; Howard Hughes Medical Institute, University of California Santa Cruz, Santa Cruz, CA; South Australian Museum, Adelaide, South Australia, Australia; Palaeogenomics Group, Department of Veterinary Sciences, Ludwig Maximilian University, Munich, Germany