
A rose may be a rose may be a rose (to paraphrase Gertrude Stein) but genetically roses (like many plant species) are wickedly complex. The genus Rosa comprises about 200 species, although only 8-20 species are thought to have contributed to modern rose cultivars. Rose genomic complexity is expressed, inter alia, in their chromosome structures, which is a genetic patchwork of sequences from a large number of cultivars, and as with many plants a significant percentage of Rosa species (about half) are polyploid. The evolutionary history of these species has involved reticulate evolution, interspecific hybridization, introgression and polyploidization, which has contributed to the genetic variability and chromosome structure. This provides broad phenotypic variability, in flower size, color, fragrance, medicinal properties, and other features selected for not (just) naturally but by generations of plant breeders. For example, one of the first hybrids, "La France," produced in Europe beginning in the 18th Century, combined "growth vigor" characteristic of European rose varieties with the propensity for current blooming characteristic of Chinese roses (as well as color and scent signatures). And this genetic complexity has made determining the rose genomic DNA sequence particularly difficult, despite its relatively small size (560 Mb).
Last spring (appropriately) a team of scientists* revealed the first compilation of the rose genome in a paper published in Nature Genetics entitled "The Rosa genome provides new insights into the domestication of modern roses." Relevant to the genetic mosaicism referenced above, these authors needed to produce a "homozygous" genotype from one of the heterozygous diploid rose progenitors, Rosa chinensis "Old Blush," in order to produce a reliable genomic rose sequence. Upon sequencing, these scientists were able to produce 7 "pseudo chromosomes" corresponding to a homozygous genome; in nature roses have 7, 14, 28, 35, or 42 chromosomes. The sequenced data suggested there are 36,377 protein-coding genes and almost 4,000 long noncoding RNA sequences in the rose genome. Transposable elements comprised almost 68% of the rose genome, with long terminal repeat retrotransposons representing about half of these sequences. Comparisons between related species permitted these researchers to identify an ancestral karyotype having 9 protochromosomes and 8,861 protogenes, with the ancestral strawberry and rose genomes were modeled into 8 protochromosomes comprising 13,060 protogenes, produced by two ancestral chromosomes fusions and one chromosome fission; the authors report that the strawberry ancestral genome experienced an extra fusion resulting in the modern genome, where the rose genome experienced one fission and two fusions, independently of strawberry.
Turning to modern rose cultivars, sequencing detected "discrete levels of variant density along the genomes of hybrid cultivars" that could "reflect different introgression histories." Three ancestral pools of diversity representing combinations of Cinnamomeae, Synstylae and Chinenses cultivar genomes could be found in various of the mosaic chromosomes detected by their sequencing results. These results then were extended to identify candidate genes that may be involved in recurrent blooming (contributed by the Rosa chinensis ancestor. One of these genetic events is "insertion of a [transposable element] in TFL1 (RoKSN), a repressor of floral transition responsive to activation by gibberellic acid," considered according to these authors as a major determinant in recurrent blooming and having arisen in Rosa chinensis cultivars. The authors also report that two candidates for s second, recently detected locus associated with recurrent blooming, as being either "transcription factor SPT [] which is known to control flowering in Arabidopsis" and "DOG1, which is known to modify flowering by acting on miR156."
Finally, these authors report "[d]ata mining of the rose genome combined with in-depth biochemical and molecular analyses of volatile organic compounds permitted identification of at least 22 biosynthetic steps in the terpene pathway that have not been characterized in the rose, two of which have not previously been characterized in other species." These studies result in a proposal that negative regulation by microRNA mi156 of repressor of anthocyanin biosynthesis (SPL9) provides a regulatory hub that "orchestrates the coordination of production of both colored anthocyanins and certain terpenes," as illustrated in Figure 3 of the paper (click on image to enlarge):
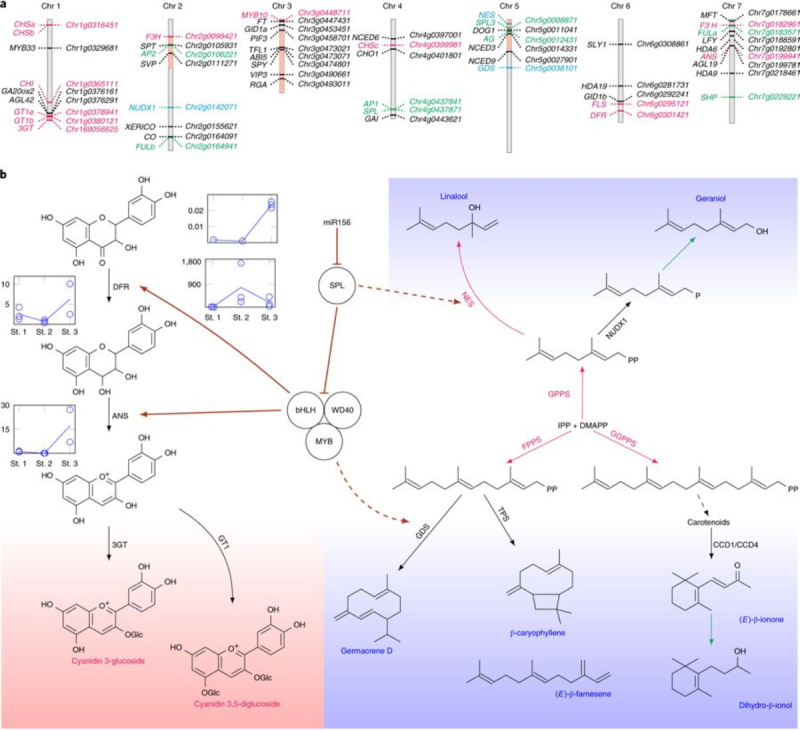
The paper concludes by noting both the scientific benefits of the study and its practical implications:
The very high-quality rose genome sequence reported in this study, combined with an expert annotation of the main pathways of interest for the rose . . . provides new insights into the genome dynamics of this woody ornamental and offer a basis to disentangle the seemingly mandatory trait associations or exclusions. Furthermore, access to candidate genes, such as those involved in abscisic acid synthesis and signaling, paves the way for improving rose quality with better water-use efficiency and increased vase life. Breeding for other characteristics such as increased resistance to pathogens should also benefit from these data and may lead to decreased use of pesticides.
The rose is one of the most commonly cultivated ornamental plants anywhere in the world, making understanding of rose genetics economically as well as scientifically important.
-
Olivier Raymond, Jérémy Just, Marion Verdenaud, Philippe Vergne, Sébastien Carrère, Judit Szécsi, Léa François, Annick Dubois, Antoine Larrieu, Lauriane Perrier, Priscilla Villand, Claudia Bardoux, Véronique Boltz, Teva Vernoux, Michiel Vandenbussche & Mohammed Bendahmane** from the Laboratoire Reproduction et Développement des Plantes, Univ Lyon, ENS de Lyon, UCB Lyon 1, CNRS, INRA, Lyon, France; Jérôme Gouzy, Hélène Badouin & Ludovic Cottret from LIPM, Université de Toulouse, INRA, CNRS, Castanet-Tolosan, France; Hélène Badouin from Univ Lyon, Université Lyon 1, CNRS, Laboratoire de Biométrie et Biologie Evolutive UMR5558, Villeurbanne, France; Marion Verdenaud, David Latrasse, Magali Perez, Adnane Boualem, Abdelhafid Bendahmane & Moussa Benhamed from the Institute of Plant Sciences Paris-Saclay (IPS2), CNRS, INRA, University Paris-Sud, University of Evry, University Paris-Diderot, Sorbonne Paris-Cite, University of Paris-Saclay, Orsay, France; Arnaud Lemainque, Arnaud Couloux, Jean-Marc Aury, Mohammed-Amin Madoui, Karine Labadie & Patrick Wincker from CEA–Institut de Biologie François Jacob, Genoscope, Evry, France; Sandrine Moja, Jean-Claude Caissard & Sylvie Baudino from Univ Lyon, UJM-Saint-Etienne, CNRS, Saint-Etienne, France; Nathalie Choisne & Hadi Quesneville from UR1164–Research Unit in Genomics-Info, INRA, Université Paris-Saclay, Versailles, France; Caroline Pont & Jérôme Salse from INRA/UBP UMR 1095 Genetics, Diversity and Ecophysiology of Cereals, Clermont-Ferrand, France; Xiaopeng Fu from Key Laboratory of Horticultural Plant Biology, College of Horticulture & Forestry Sciences, Huazhong Agricultural University, Wuhan, China; Shu-Hua Yang from Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China; Florence Piola from Univ Lyon, Université Claude Bernard Lyon 1, CNRS, ENTPE, UMR5023 LEHNA, Villeurbanne, France; Benjamin Govetto, Yoan Labrousse & Manuel Le Bris from Aix Marseille Université, Avignon Université, CNRS, IRD, IMBE, Institut Méditerranéen de Biodiversité et d'Ecologie, Marseille, France; Céline Lopez-Roques from INRA, US 1426, GeT-PlaGe, Genotoul, Castanet-Tolosan, France; Pascal Heitzler from Institut de Biologie Moléculaire des Plantes, CNRS, UPR 2357, Strasbourg, France; Chang Liu from Center for Molecular Biology (ZMBP), University of Tübingen, Tübingen, Germany; and Patrick Wincker from CNRS, Université d'Evry, UMR 8030, Evry, France.
** Corresponding author