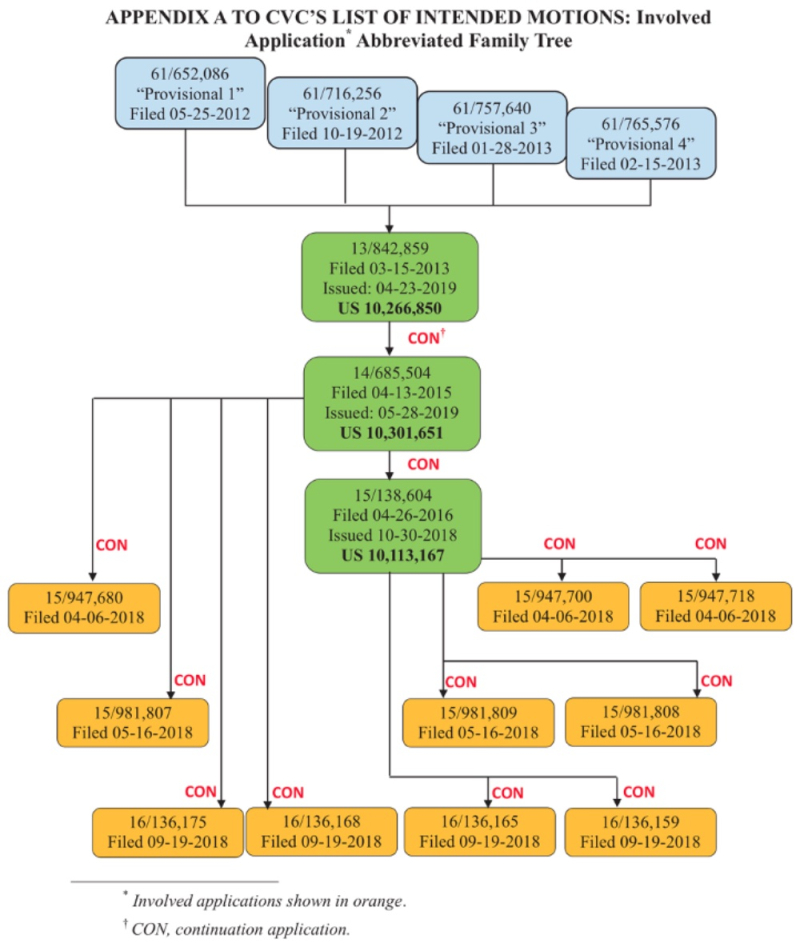
In Interference No. 106,115 between Senior Party the Broad Institute (joined by Harvard University and MIT) and Junior Party the University of California, Berkeley; the University of Vienna; and Emmanuelle Charpentier (collectively, "CVC"), the Patent Trial and Appeal Board granted CVC's Preliminary Motion for benefit of priority to U.S. Provisional Application No. 61/757,640, filed January 28, 2013 ("Provisional 3"), pursuant to 37 C.F.R. §§ 41.121(a)(1)(ii) and 41.208(a)(3) and Standing Order ¶ 208.4.1. In Interference No. 106,127, Senior Party ToolGen filed its Substantive Preliminary Motion No. 1 to deny CVC priority benefit to the '640 provisional application.
The relationships between the patents and applications in the '127 interference are set forth in this chart (filed in CVC's earlier preliminary motion in the '115 Interference):
ToolGen (stating that Broad did not substantively challenge CVC's assertion of this priority) challenges CVC's entitlement to priority benefit to the P3 provisional here on the grounds that it did not disclose "successful cleavage of DNA within eukaryotic cells, nor does it otherwise show a constructive reduction to practice of an embodiment within Count 1."
ToolGen's argument depends on three assertions. First, the brief identifies a single Example (Example 2) in the P3 provisional application directed towards purported eukaryotic cell embodiments of CRISPR gene editing (which, if it provided an adequate description of even a single enabled embodiment would be sufficient to satisfy the requirement for priority). Second, ToolGen asserts that the Example (and Figures 38B and 36E related thereto) do not provide such an adequate description because "Figure 38B shows alleged cleavage bands at positions where there should be none and in some instances no bands where there should be bands." Similarly, ToolGen asserts that Figure 36E "independently contains so many unexplained bands as to make it so shaky and unreliable that a [person of ordinary skill in the art or] POSA would not view it as showing possession of an embodiment within Count 1." Taken together, ToolGen argues that "the Figures cannot be evidence that discloses successful cleavage to a POSA in eukaryotes, and the applicants' failure to sequence the resulting products further cements this conclusion." Third, ToolGen argues that "applicants lysed the cells before DNA extraction such that they left the Cas9 protein active to cleave DNA outside of intact cells, as opposed to within the eukaryotic cell as required by Count 1" and "a POSA would understand that any cleavage shown in the gel results cannot confirm that cleavage occurred within eukaryotic cells." Like U.S. provisional applications 61/652,086 ("P1") and 61/716,56 ("P2") (for which the Board refused to recognize priority for failure to disclose an adequate description of even a single enabled embodiment of CRISPR in a eukaryotic cell), ToolGen argues that the P3 provisional application is similarly deficient and the Board should deny CVC the accorded priority benefit.
After setting forth the statutory requirements for a party to be accorded benefit of priority and discussing the level of ordinary skill in the art, ToolGen sets forth its analysis of the deficiencies in the P3 disclosure synopsized above. Supported by expert testimony (Dr. Turchi), ToolGen argues that a POSA at the time of the invention would not have considered the P3 disclosure to satisfy these requirements. Like Broad, ToolGen argues that "adapting the native prokaryotic CRISPR-Cas9 system to cleave DNA within eukaryotic cells was highly unpredictable." Like Broad before it ToolGen enumerates the many differences between the prokaryotic milieu where CRISPR was expressed natively and eukaryotic cells (the existence of the nucleus, packaging genomic DNA in chromatin, the intracellular components for gene expression in eukaryotic cells, and the resulting uncertainty ToolGen alleges arises as a consequence regarding whether the CRISPR-Cas9 system could be adapted for use in these cell types. (An uncertainty ToolGen notes that the Board acknowledged supported by the now infamous, improvident statements by CVC's inventors.)
Besides these somewhat theoretical obstacles (which, had CRISPR been readily reduced to practice would not have their current asserted significance), ToolGen critically reviews what it contends are deficiencies in the disclosure of Example 2 as illustrated by Figures 38B and 36E (as these deficiencies were attested to by ToolGen's expert witness). This analysis was set out graphically to show the expected DNA fragments and sequences that the skilled worker would expect and need to observe to conclude successful CRISPR gene editing in the HEK293T human embryonic kidney cells used in the experiments disclosed in Example 2 of the P3 provisional application:
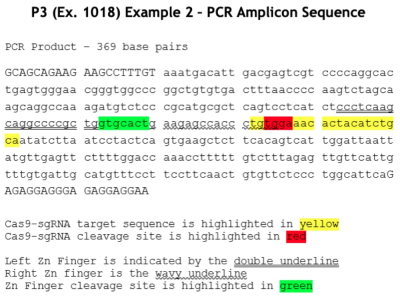
The brief then sets out the bands produced by successful CRISPR according to ToolGen's expert:
• A 369 bp PCR product band because not all of the DNA would have been cleaved. Any larger bands would be unexpected because the PCR product should contain only 369 bp DNA fragments in all lanes.
• Approximately 183 and 186 bp Cas9-sgRNA cleavage bands in the lanes with both Cas9 and sgRNA.
• Approximately 162-169 and 200-207 bp ZFN cleavage bands in the ZFN positive control lane.
Instead of these results, the brief (relying of ToolGen's expert) asserts that:
If the Figure 38B and 36E gels showed successful DNA cleavage at the target site, a POSA would expect the data in the gels to be consistent with the above. They are not. Rather, Figure 38B contains bands that differ significantly from the bands that a POSA would expect if cleavage had been successful. Figure 36E shows results inconsistent with Figure 38B, and also contains so many unexplained bands as to render the data unreliable—and a POSA would view it as insufficient to demonstrate possession [citations to exhibits omitted].
The brief further sets forth an "annotated" version of Figure 38B to illustrate its purported deficiencies:
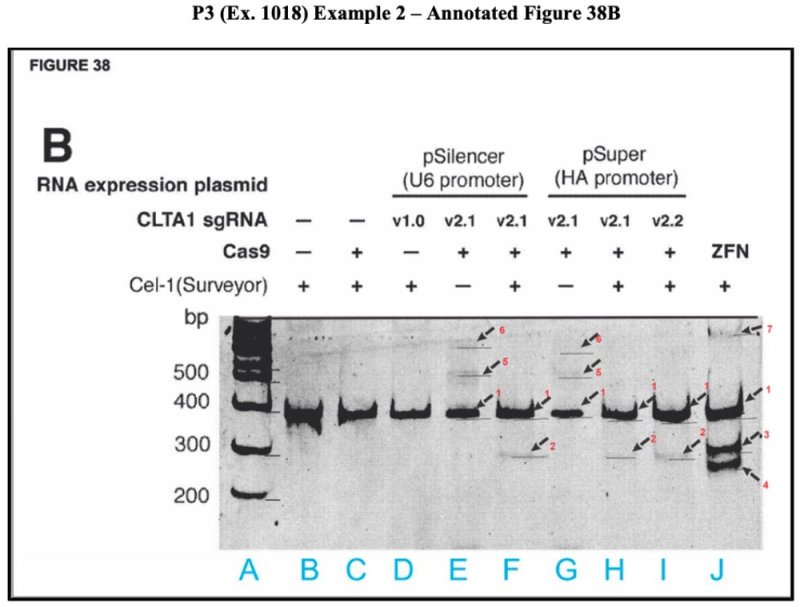
ToolGen's expert's analysis is set forth in the brief as follows (citations to the exhibits omitted):
The ~378 bp PCR band (band 1) is consistent with the predicted 369 bp full-length PCR band. In contrast, the digestion product (band 2)—which the applicants claim is a CRISPR-Cas9 cleavage band—is not the expected size in any of the Cas9-sgRNA lanes (Lanes F, H, and I). As Dr. Turchi explains, the supposed cleavage band in Figure 38B measures ~280 bp (band 2)—almost 100 bp longer than the expected ~185 bp. There is no band at all in the expected ~185 bp range—which should have been identifiable at slightly below the 200 bp control ladder marker. It is also irreconcilable with the sizes of the other bands.
If the ~280 bp band (band 2) was a digestion band from the ~378 bp PCR band (band 1), there is an unaccounted for ~98 bp DNA fragment, making it unlikely that the ~280 bp band is an actual cleavage band. Ex. 1410 88; F23. There are also several unexpected bands—larger than the PCR band—of unknown origin that disappear upon digestion (bands 5 and 6). Therefore, it is possible that the alleged cleavage band (band 2) comes from one of the larger, digestion sensitive bands and not the PCR band.
Accordingly, because "applicants nowhere address the irregularities in Figure 38B or provide a POSA with any comfort that they possessed an embodiment within Count 1" and thus "a POSA at the time P3 was filed would find Figure 38B to be unreliable and would have disregarded it in its entirety."
The brief then sets forth a similar analysis of the experimental results shown in Figure 36E. The bands found in this Figure should be the same as those in Figure 38B, according to ToolGen's expert, but it does not. One deficiency is the presence of additional bands; as stated in the brief, "[a]lmost every lane has more than one unexpected band, making it impossible to determine the origin of each digestion product band, which P3 does not attempt to explain." As with Figure 38B, ToolGen's expert prepared an annotated version of this Figure:
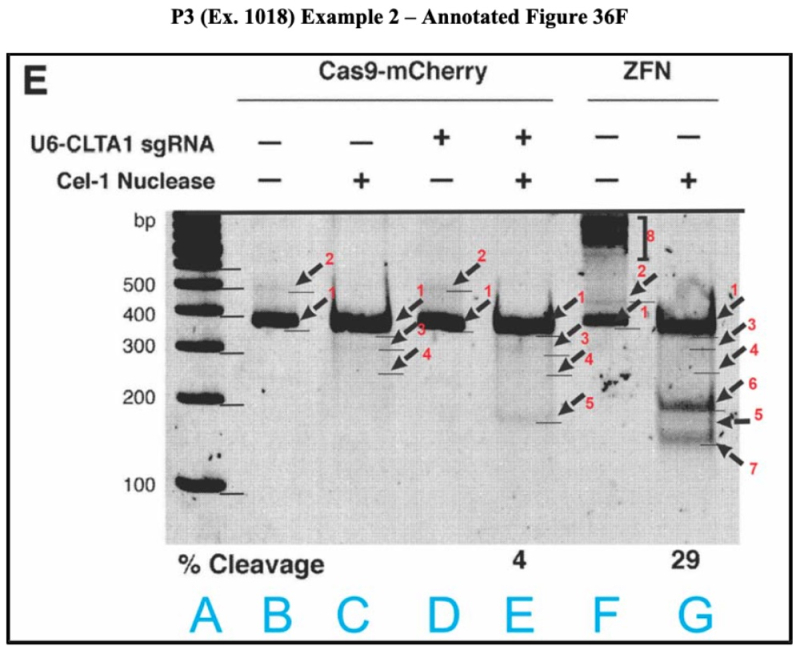
As explained by ToolGen's expert:
The PCR band (band 1) measures ~352 bp, consistent with its expected size. The applicants rely on a single ~173 bp band (band 5) in Lane E to indicate CRISPR10 Cas9 cleavage at the target site. However, the alleged cleavage band (band 5) could just as easily be a digestion product of the unexpected ~477 bp band (band 2) in Lanes B, D, and F that disappear upon digestion in the Surveyor assay Lanes E and G. This is fully consistent with the presence of an unexpected digestion band at ~297/312 bp (band 3) also in Lanes E and G, because the unexpected ~477 bp band (band 2) could have been digested into the ~173 and ~297/312 bp digestion product bands (bands 5 and 3, 4 respectively). And there is yet another unexpected digestion product band at ~252 bp (band 4) in Lanes E and G, which could reflect the unexpected ~477 bp band (band 2) being digested into two equal DNA fragments. These anomalies would leave the POSA unconvinced of the applicants' possession of an embodiment within Count 1.
As Dr. Turchi notes, a digestion band is visible in Lane G—between the two ZFN cleavage bands—of similar size and intensity as the purported cleavage band (band 5) in Lane E. This digestion band is significant because it would further lead a POSA to conclude that band 5 in Lane E is simply a digestion product of one of the larger unexpected band (band 2) in the undigested lanes (Lanes, B, D, and F), and not indicative of successful Cas9 cleavage at the target site.
From this analysis ToolGen's expert opines that the evidence in Figure 36E is "inconclusive" and "riddled with anomalies and uncertainties" sufficiently inconsistent with CVC's assertion of successful practice of CRISPR-Cas9 in eukaryotic cells that these assertions would not be believed by a person of ordinary skill in the art.
In view of the asserted poor quality of these experimental results, ToolGen further asserts that the skilled worker "next have performed sequencing on the PCR product to confirm and characterize any potential cleavage results." Yet, as the brief notes, "no such work was reported in [the] P3 [provisional application]."
As the final asserted defect in the evidentiary support for CVC's purported reduction to practice of CRISPR in eukaryotic cells, the brief then sets forth its argument that the way the cells were lysed after introduction of CRISPR-Cas9 could have permitted any observed cleavage to have occurred after lysis and thus fall outside the scope of the invention as defined by the Count. This is illustrated in annotated Figure 37A:
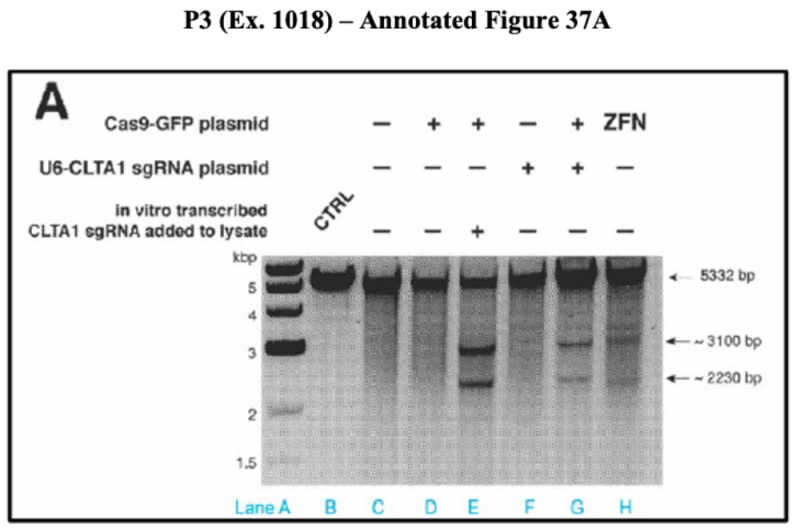
As set forth in the brief, "the presence of two bands indicates to a POSA that the plasmid was successfully cleaved extracellularly at the predicted Cas9-sgRNA target site and the donor plasmid restriction enzyme site" (emphasis added). As a consequence, the brief goes on to say that "[t]he results shown in Figure 37A undermine any arguable cleavage results in Figures 38B and 36E because they show that extracellular cleavage occurred in cell lysates prepared using the same Cas9-preserving protocol that was used to prepare the cell lysate in Figure 38B and 36E [emphasis in the brief]. Because the applicants preserved the activity of Cas9 in the lysate, any alleged cleavage shown in Figures 38B or 36E could have occurred outside the cells, in the lysate."
In addition, the relative cleavage efficiencies between the ZFN positive control and the purported CRISPR-Cas9 lanes further support the conclusion that such CRISPR-Cas9-mediated cleavage occurred in vitro in the lysis buffer and thus does not support reduction to practice of eukaryotic cell embodiments of CRISPR gene editing by CVC in the P3 provisional priority document.
ToolGen thus asks the Board to rule that CVC is not entitled to the benefit of priority of the P3 provisional application.